Tutorial
This tutorial will guide you through the steps of using the MariClus database to explore the natural products and gene clusters of marine species. You will learn how to:
- Search for a specific molecule or gene cluster
- View the details and evidence of a gene cluster
- Examine the antiSMASH results of a gene cluster
- Access additional information on PubChem
Note:
Mariclus uses different styles of underlined text. An overview is given below:
- Black underlined text means that the text can be clicked to expand or collapse additional information. It can also link to information on the same page (like the table of contents on this page)
- Blue underlined text means that the text is a hyperlink to another (internal or external) page
- Dashed lines means that the text contains additional information when hovering over it
Step 1: Search for a molecule or gene cluster
To start, go to the 'Molecules' section of MariClus and enter the name of the molecule you are interested in. For example, if you want to find out more about abyssomicin, a unique antimicrobial that inhibits folate synthesis, type 'abyssomicin' in the search box.
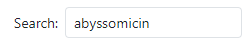
This will retrieve a list of marine species and gene clusters associated with abyssomicin. The list is ranked according to a cumulative BLAST score of the genes present in the identified cluster compared to the known abyssomicin C gene cluster of Micromonospora maris AB-18-032, which is available on the Minimum Information about a Biosynthetic Gene cluster (MIBiG) database with accession number BGC0000001. The higher the score, the more likely the cluster is related to abyssomicin.
See the example search live: abyssomicin
Step 2: Use the advanced filtering options
Either inside the species, clusters or molecules page, several advanced filtering options can be applied. On the species and clusters page, an advanced species filter can be used. To filter on multiple species, click on the “Show advanced filtering options” button to display the search box. In this box, enter the species of interest, separated by commas (see the image below). Conveniently, the full name of the species does not have to be provided. For example, if you are interested in the genera Candidatus, Polaribacter and Streptomyces, enter these genera, separated by commas.
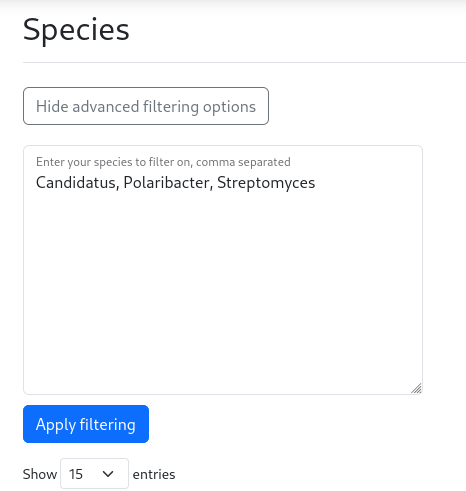
See the example search live here
On the molecules page, there is an advanced molecular mass filter and product name filter. Again, when clicking on the “Show advanced filtering options”, the filter options will be displayed. To use these, enter your prefered molecular mass range and additionaly, a list of products of interest can be provided.
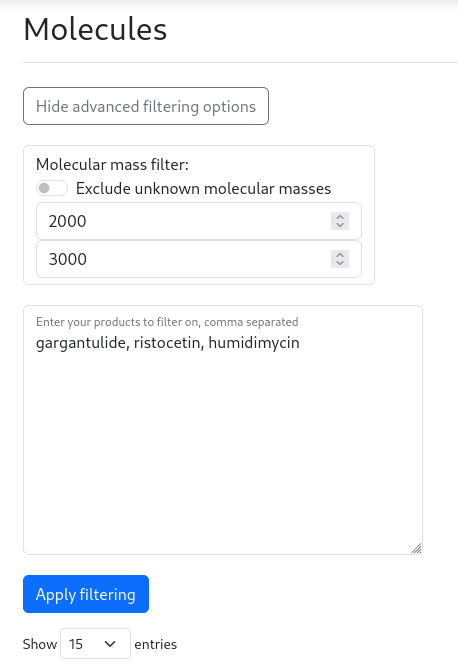
See the example search live here
Step 3: View the details and evidence of a gene cluster
To view more details about a gene cluster, click on the 'Details' icon next to the species name. This will take you to the respective details page, where you get general information about the species as well as an overview of all detected gene clusters.
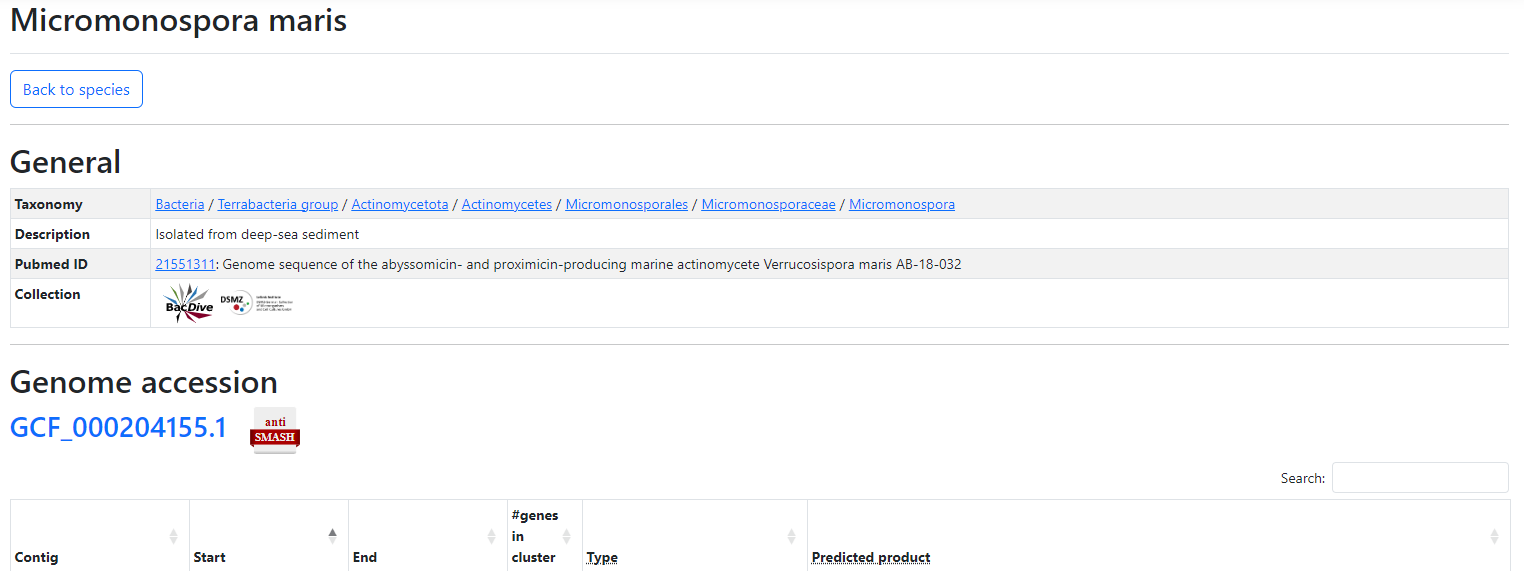
Furthermore, if you came directly from the molecules page, the page scrolls down to the section of interest. These sections contain information about the genomic region, the start and ending positions on this region, number of genes in this cluster, and the predicted product type. The molecule of interest is highlighted in yellow.

To see how the gene cluster was predicted by antiSMASH, click on the 'Evidence' icon next to the product type. This will show you a table with information on each gene in the cluster, such as its name, function, length, and BLAST score against the known abyssomicin C gene cluster. You can also see which genes are correlated to the MIBiG reference cluster and which ones are unique or different.
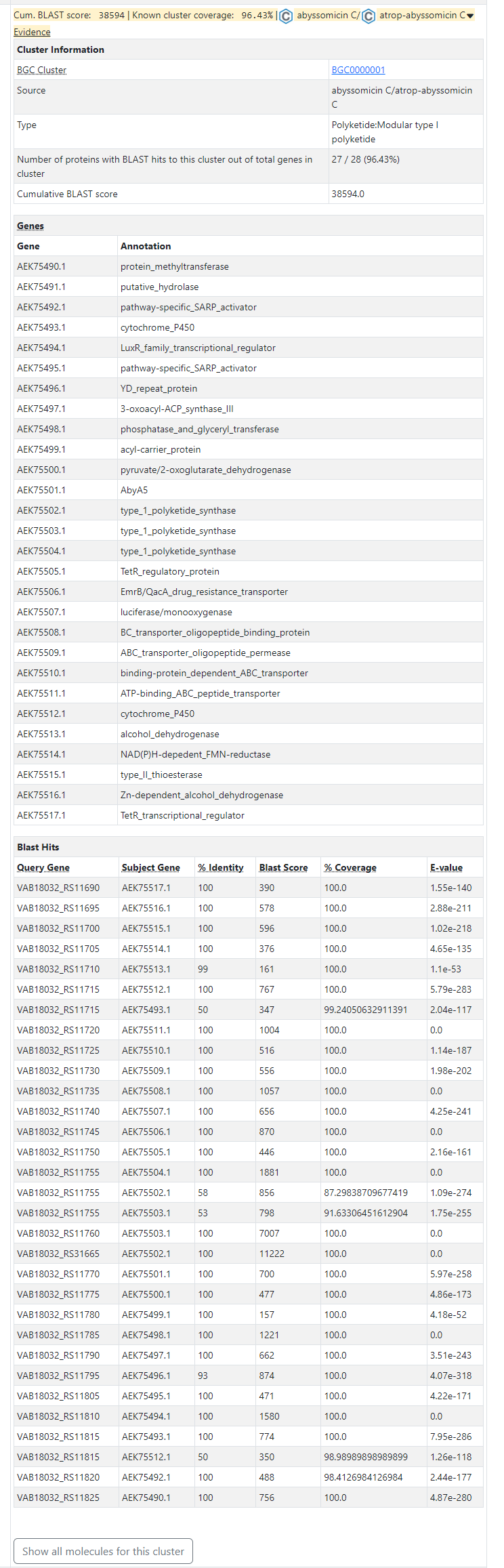
See the example details page live: Micromonospora maris
Step 4: Examine the antiSMASH results of a gene cluster
MariClus provides a detailed and transparent report on the antiSMASH natural product prediction by gene clusters. You can use this feature to examine the genomic features and functions of a gene cluster of interest. For example, if you want to learn more about the abyssomicin gene cluster of Micromonospora maris, you can click on the antiSMASH icon next to the cluster.
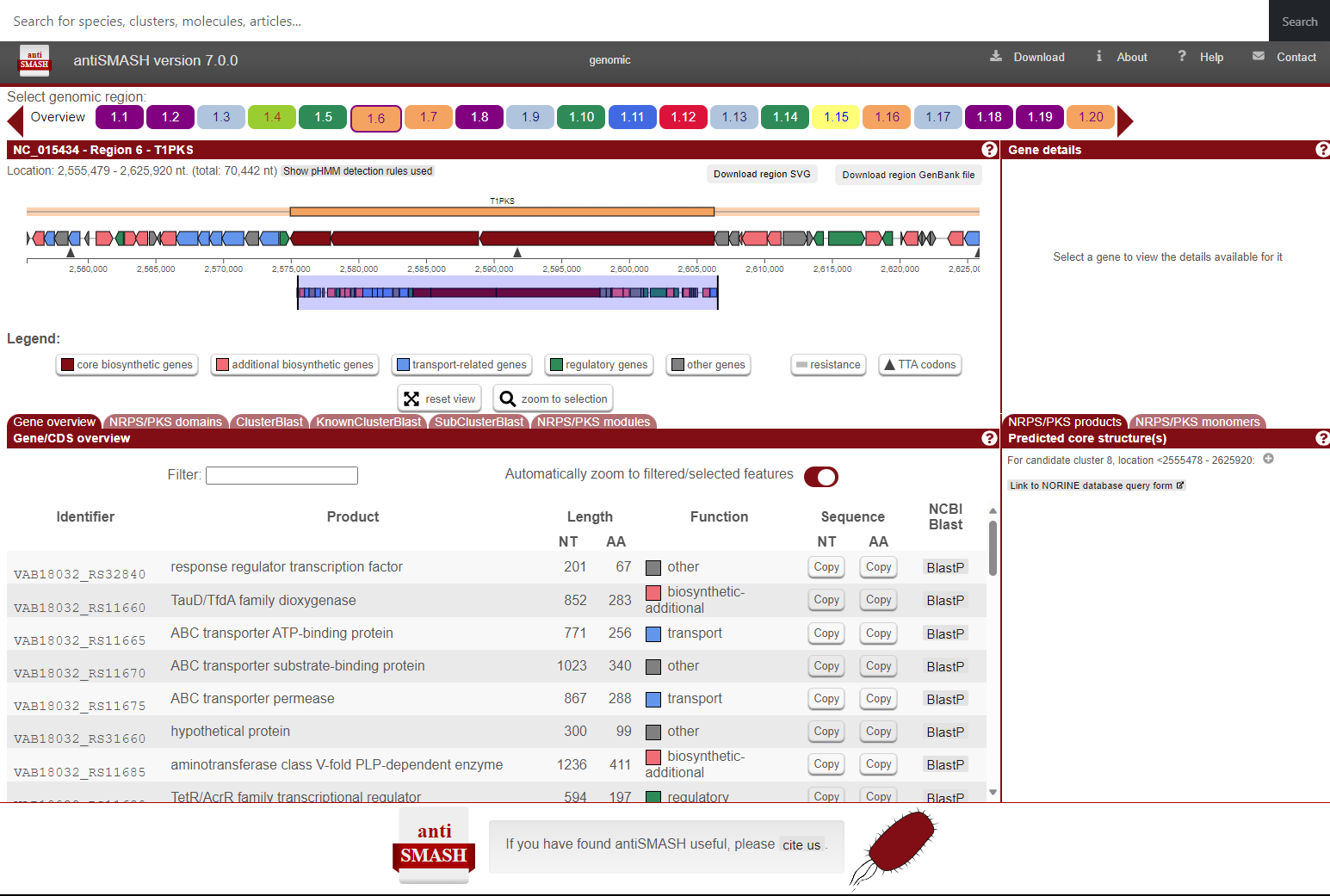
This will take you to the antiSMASH gene cluster webpage, which contains all features normally available when using antiSMASH as a standalone tool. You can see the genomic region, the coding sequences, the predicted product type, and the annotations for each gene. You can also use the KnownCluster-Blast feature to compare the gene cluster with known gene clusters from other species or databases.
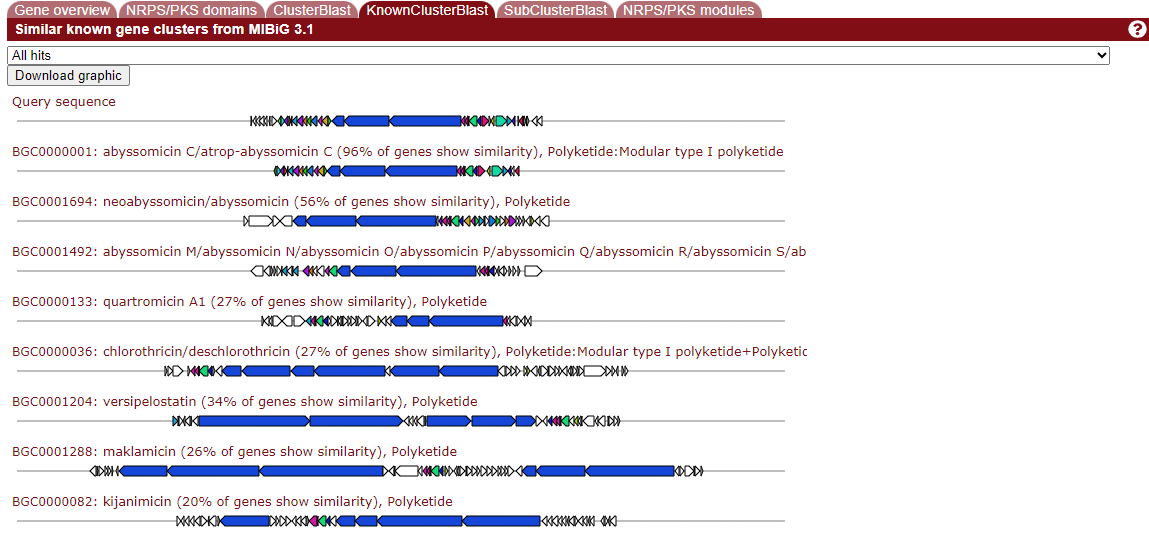
In this way, MariClus enables enhanced genome mining as it is not limited to comparing a certain gene cluster to only 10 known ones, but rather to all available ones in the database.
See the example antiSMASH page live: Micromonospora maris abyssomicin C
Step 5: Access additional information on PubChem
As marine natural products are a valuable source of pharmaceutical drugs, they have been researched extensively and reported in various sources. MariClus therefore links the predicted compounds produced by marine species to their PubChem page, where you can find additional information on biochemical pathways, assays, literature, and patents. For example, the marine natural product salinosporamide A was discovered in 2003 from marine Salinispora species and has since been commercialized as the proteasome inhibiting anti-cancer drug Marizomib. Consequently, many papers and patent applications have been published.
The PubChem icon next to salinosporamide A of Salinispora tropica in the MariClus database directly links to PubChem, where you can find an overview of (at the time of writing) 91 substances, 21 related compounds, 3 biochemical pathways, 137 assays, 103 manuscripts and 100 patents.
See the example PubChem page live: salinosporamide A
Final remarks
We hope this tutorial was helpful and that you enjoy using MariClus. If you have any questions or feedback, please contact us via the contact form.